ARCHIVED: Use GROMACS on Big Red II at IU
On this page:
- Overview
- Set up your user environment
- Prepare a GPU-accelerated batch job script
- Performance benefits of using GPU-accelerated GROMACS
- Prepare a CPU-only batch job script
- Submit and monitor your job
- Get help
Big Red II was retired from service on December 15, 2019; for more, see ARCHIVED: About Big Red II at Indiana University (Retired).
Overview
GROMACS (Groningen Machine for Chemical Simulations) is a full-featured molecular dynamics simulation suite designed primarily for studying proteins, lipids, and polymers, but useful for analyzing a wide variety of chemical and biological research questions, as well. Originally developed at the University of Groningen, GROMACS is open source software maintained by contributors at universities and research centers in Sweden, Germany, and the US. GROMACS is freely available under the GNU Lesser General Public License (LGPL). Serial and MPI versions are available with 64-bit addressing. Starting with version 4.6, GROMACS includes native, CUDA-based GPU acceleration support for NVIDIA Fermi and Kepler GPUs.
At Indiana University, Big Red II has single precision and double precision versions of GROMACS for running parallel jobs on compute nodes in the Extreme Scalability Mode (ESM) execution environment. GPU-accelerated versions also are available for running parallel jobs on Big Red II's hybrid CPU/GPU nodes. The GPU-accelerated versions routinely run twice as fast as the CPU versions. To take advantage of GPU acceleration you must use a GPU-accelerated version of GROMACS, include the NVIDIA CUDA Toolkit in your user environment, and specify the GPU queue in your job script.
Set up your user environment
To add GROMACS to your Big Red II user environment, you first must add the GNU programming environment and the FFTW (Fastest Fourier Transforms in the West) subroutine library. To use a GPU-accelerated version of GROMACS, you also must add the NVIDIA CUDA Toolkit.
GMXRC script (which identifies the shell you're running and sets GROMACS environment variables accordingly), on Big Red II all necessary user environment changes are handled automatically when you load any of the available GROMACS modules.
Use Module commands to add the necessary packages to your user environment:
- Verify that the
PrgEnv-gnumodule is added to your user environment; on the command line, entermodule listto determine which modules are currently loaded.If another programming environment module (for example,
PrgEnv-cray) is loaded, use themodule swapcommand to replace it with thePrgEnv-gnumodule:module swap PrgEnv-cray PrgEnv-gnu
- If the FFTW library is not listed among the currently loaded modules, load the default
fftwmodule; on the command line, enter:module load fftw
- Check which versions of GROMACS are available; on the command line, enter:
module avail gromacs
Note:Either of the single precision or double precision versions are suitable for running GROMACS jobs on Big Red II's compute nodes. Although these versions will run on the hybrid CPU/GPU nodes, they cannot take advantage of GPU acceleration. To run GPU-accelerated jobs on Big Red II's hybrid CPU/GPU nodes, choose a GPU-accelerated version. - To load the version of your choice, on the command line, enter:
module load gromacs/version_path
Replace
version_pathwith the path corresponding to version you want to load; for example:- To load the double precision version, enter:
module load gromacs/gnu/double/4.6.2
- To load the GPU-accelerated version, enter:
module load cudatoolkit module load gromacs/gnu/gpu/4.6.5
- To load the double precision version, enter:
For example, to automatically load upon login the modules needed to run a GPU-accelerated GROMACS job, add the following lines to your ~/.modules file:
module swap PrgEnv-cray PrgEnv-gnu module load fftw module load cudatoolkit module load gromacs/gnu/gpu/4.6.5
Prepare a GPU-accelerated batch job script
To run a parallel GROMACS job in batch mode on the hybrid CPU/GPU nodes in Big Red II's native ESM execution environment, prepare a TORQUE job script (for example, ~/work_directory/my_job_script.pbs) that specifies the application you want to run, sets the resource requirements and other parameters appropriate for your job, and invokes the aprun command to properly launch your application.
On Big Red II:
- To invoke the MPI version of a GROMACS command, append the
_mpisuffix to its name (for example,mdrun_mpi). - To invoke the double precision MPI version of a GROMACS command, append the
_mpi_dsuffix to its name (for example,mdrun_mpi_d).
The following sample script executes the MPI version of mdrun on four Big Red II CPU/GPU nodes in the ESM execution environment:
#!/bin/bash #PBS -l nodes=4:ppn=16,walltime=3:00:00 #PBS -q gpu #PBS -o out.log #PBS -e err.log cd $PBS_O_WORKDIR aprun -n 4 -N 1 mdrun_mpi <mdrun_mpi_options>
In the sample script above:
- The
-q gpuTORQUE directive routes the job to thegpuqueue. - The
cd $PBS_O_WORKDIRline changes to the directory from which the job was submitted and where the input files are. - If you did not previously add the required modules to your user environment (as described in the preceding section), you must include the necessary
module loadcommands in your script before invoking theapruncommand. - When invoking
aprunon the CPU/GPU nodes, the-nargument specifies the total number of nodes (not the total number of processing elements), and the-Nargument specifies the number of GPUs per node, which is one (for example,-N 1). - Replace
<mdrun_mpi_options>withmdrun_mpioptions indicating your input and output files, and other parameters for controlling your simulation; for descriptions of available options, see themdrunmanual page (man mdrun).
Performance benefits of using GPU-accelerated GROMACS
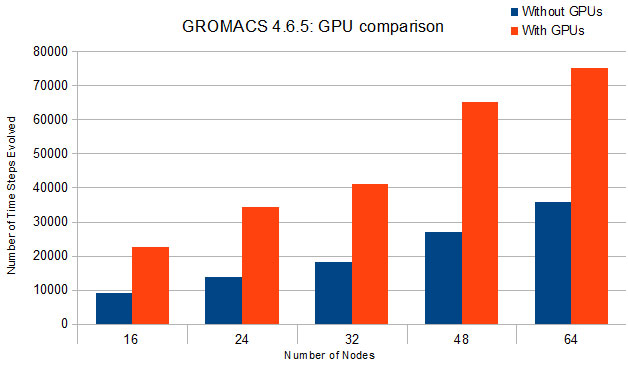
The following graph compares the results of test simulations run using a CPU version of GROMACS with results of identical simulations run using a GPU-accelerated version. The test simulation involved more than 14 million atoms. Each run had identical simulation parameters and runtimes, and all simulations were performed on the hybrid CPU/GPU nodes. The data values reported represent the number of time steps evolved during the simulations (larger values indicate better performance). On each run, the GPU-accelerated version out-performed the CPU version by a significant margin.
Prepare a CPU-only batch job script
To run a parallel GROMACS job in batch mode on the compute nodes in Big Red II's native ESM execution environment, prepare a TORQUE job script (for example, ~/work_directory/my_job_script.pbs) that specifies the application you want to run, sets the resource requirements and other parameters appropriate for your job, and invokes the aprun command to properly launch your
application.
On Big Red II:
- To invoke the MPI version of a GROMACS command, append the
_mpisuffix to its name (for example,mdrun_mpi). - To invoke the double precision MPI version of a GROMACS command, append the
_mpi_dsuffix to its name (for example,mdrun_mpi_d).
The following sample script executes the MPI version of mdrun on two Big Red II compute nodes in the ESM execution environment:
#!/bin/bash #PBS -l nodes=2:ppn=32,walltime=6:00:00 #PBS -q cpu #PBS -o out.log #PBS -e err.log cd $PBS_O_WORKDIR aprun -n 64 mdrun_mpi <mdrun_mpi_options>
In the sample script above:
- The
-q cpuTORQUE directive routes the job to thecpuqueue. - The
cd $PBS_O_WORKDIRline changes to the directory from which the job was submitted and where the input files are. - If you did not previously add the required modules to your user environment (as described in the preceding section), you must include the necessary
module loadcommands in your script before invoking theapruncommand. - Replace
<mdrun_mpi_options>withmdrun_mpioptions indicating your input and output files, and other parameters for controlling your simulation; for descriptions of available options, see themdrunmanual page (man mdrun).
Submit and monitor your job
To submit your job script (such as
~/work_directory/my_job_script.pbs), use the TORQUE
qsub command; for example, on the command line,
enter:
qsub [options] ~/work_directory/my_job_script.pbs
For a full description of the qsub command and
available options, see its manual page.
To monitor the status of your job, use any of the following methods:
- Use the TORQUE
qstatcommand; on the command line, enter (replaceusernamewith the IU username you used to submit the job):qstat -u username
For a full description of the
qstatcommand and available options, see its manual page. - Use the Moab
checkjobcommand; on the command line enter (replacejob_idwith the ID number assigned to your job):checkjob job_id
For a full description of the
checkjobcommand and available options, see its manual page.
Get help
To learn more about GROMACS:
- See the tutorials and online documentation on the GROMACS website.
- Download the GROMACS Version 4.6.5 user manual.
- Consult the GROMACS manual page for brief descriptions of the individual GROMACS commands. Each GROMACS command has its own manual page that lists its options and their functions.
If you need help or have questions about running GROMACS jobs on Big Red II, contact the UITS Research Applications and Deep Learning team. If you have system-specific questions about Big Red II, or if you encounter problems running batch jobs, contact the UITS High Performance Systems group.
This is document besc in the Knowledge Base.
Last modified on 2023-04-21 16:59:33.